Cross-type Biomedical Named Entity Recognition with Deep Multi-task Learning
Xuan Wang, Yu Zhang, Xiang Ren, Yuhao Zhang, Marinka Zitnik, Jingbo Shang, Curtis Langlotz and Jiawei Han. Bioinformatics, 2018.
Biomedical Named Entity Recognition (BioNER)
Biomedical named entity recognition (BioNER) is one of the most fundamental task in biomedical text mining that aims to automatically recognize and classify biomedical entities (e.g., genes, proteins, chemicals and diseases) from text. State-of-the-art BioNER systems often require handcrafted features (e.g., capitalization, prefix and suffix) to be specifically designed for each entity type. This feature generation process takes the majority of time and cost in developing a BioNER system, and leads to highly specialized systems that cannot be directly used to recognize new types of entities. Recent NER studies consider neural network models to automatically generate quality features. However, these models have millions of parameters and require very large datasets to reliably estimate the parameters. This poses a major challenge for biomedicine, where datasets of this scale are expensive and slow to create and thus neural network models cannot realize their potential. Although neural network models can outperform traditional sequence labeling models (e.g., CRF models), they are still outperfomed by handcrafted feature-based systems in multiple domains.
Our Framework
We propose a new multi-task learning framework using character-level neural models for BioNER. The proposed framework, despite being simple and not requiring any feature engineering, achieves excellent benchmark performance. Our multi-task model is built upon a single-task neural network model (Liu et al., 2018). In particular, we consider a BiLSTM-CRF model with an additional context-dependent BiLSTM layer for modeling character sequences. A prominent advantage of our multi-task model is that inputs from different datasets can efficiently share both character- and word-level representations, by reusing parameters in the corresponding BiLSTM units.
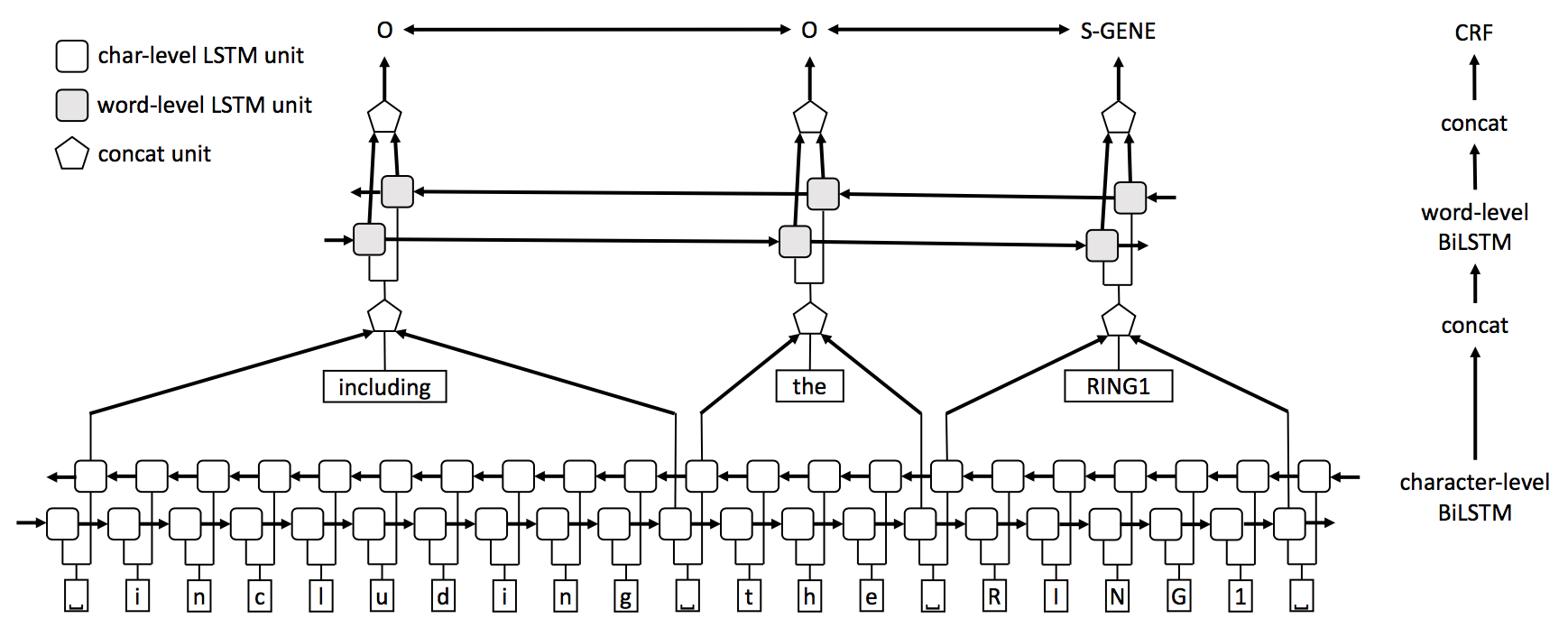
Single-task model (STM): Our single-task model consists of three layers (Figure 1). In the first layer, a BiLSTM network is used to model the character sequence of the input sentence. We use character embedding vectors as input to the network. Hidden state vectors at the word boundaries of this character-level BiLSTM are then selected and concatenated with word embedding vectors to form word representations. Next, these word representation vectors are fed into a word-level BiLSTM layer (i.e., the upper BiLSTM layer in Figure 1). Lastly, output of this word-level BiLSTM is fed into the a CRF layer for label prediction.
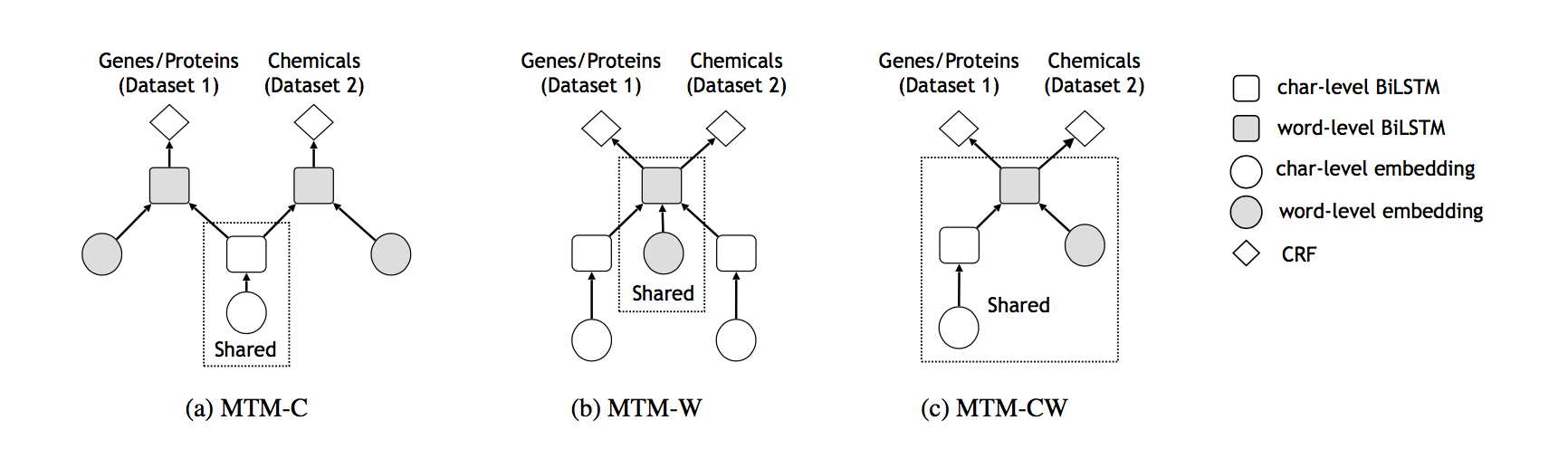
Multi-task models (MTMs): We propose three different multi-task models (Figure 2). These three models differ in which part of the model parameters are shared across multiple datasets.
MTM-C: the character-level BiLSTM parameters are shared across multiple datasets.
MTM-W: the word-level BiLSTM parameters are shared across multiple datasets.
MTM-CW: both the character-level and the word-level BiLSTM parameters are shared across multiple datasets.
Performance
We compare our model with recent state-of-the-art models on the five benchmark BioNER datasets. We use F1 score as the evaluation metric.
| Model | BC2GM | BC4CHEMD | BC5CDR | NCBI-disease | JNLPBA |
|---|---|---|---|---|---|
| Dataset Benchmark | - | 88.06 | 86.76 | 82.90 | 72.55 |
| Crichton et al. 2016 | 73.17 | 83.02 | 83.90 | 80.37 | 70.09 |
| Lample et al. 2016 | 80.51 | 87.74 | 86.92 | 85.80 | 73.48 |
| Ma and Hovy 2016 | 78.48 | 86.84 | 86.65 | 82.62 | 72.68 |
| Liu et al. 2018 | 80.00 | 88.75 | 86.96 | 83.92 | 72.17 |
| MTM-CW | 80.74 | 89.37 | 88.78 | 86.14 | 73.52 |
We also compare our single-task model (STM) and multi-task model (MTM-CW) with the baseline multi-task neural network model by Crinchton et al. on the same 15 datasets used by Crinchton et al. We use F1 score as the evaluation metric.
| Dataset | Crichton et al. 2016 | STM | MTM-CW | Performance Improvement |
|---|---|---|---|---|
| AnatEM | 82.21 | 85.30 | 86.04 | +4.19 |
| BC2GM | 73.17 | 80.00 | 78.86 | +5.69 |
| BC4CHEMD | 83.02 | 88.75 | 88.83 | +5.81 |
| BC5CDR | 83.90 | 86.96 | 88.14 | +4.24 |
| BioNLP09 | 84.20 | 84.22 | 88.08 | +3.88 |
| BioNLP11EPI | 78.86 | 77.67 | 83.18 | +4.32 |
| BioNLP11ID | 81.73 | 74.60 | 83.26 | +1.53 |
| BioNLP13CG | 78.90 | 81.84 | 82.48 | +3.58 |
| BioNLP13GE | 78.58 | 69.30 | 79.87 | +1.29 |
| BioNLP13PC | 81.92 | 85.46 | 88.46 | +6.54 |
| CRAFT | 79.56 | 81.20 | 82.89 | +3.33 |
| Ex-PTM | 74.90 | 67.66 | 80.19 | +5.29 |
| JNLPBA | 70.09 | 72.17 | 72.21 | +2.12 |
| Linnaeus | 84.04 | 86.94 | 88.88 | +4.84 |
| NCBI-Disease | 80.37 | 83.92 | 85.54 | +5.17 |
Note: Since all the datasets are trained together for MTM-CW, the performance on each dataset is slightly different when trained on 15 datasets compared with trained on 5 datasets (shown above).
Resources
Codes and datasets have been uploaded to Github.
Reference
If you find the code and datasets useful, please cite the following paper:
@article{doi:10.1093/bioinformatics/bty869,
author = {Wang, Xuan and Zhang, Yu and Ren, Xiang and Zhang, Yuhao and Zitnik, Marinka and Shang, Jingbo and Langlotz, Curtis and Han, Jiawei},
title = {Cross-type Biomedical Named Entity Recognition with Deep Multi-Task Learning},
journal = {Bioinformatics},
volume = {},
number = {},
pages = {bty869},
year = {2018},
doi = {10.1093/bioinformatics/bty869},
URL = {http://dx.doi.org/10.1093/bioinformatics/bty869},
eprint = {/oup/backfile/content_public/journal/bioinformatics/pap/10.1093_bioinformatics_bty869/1/bty869.pdf}
}